Background
Background.RmdMotivation
Microbial explicit carbon models include process-based representations of microbial activity in soil carbon models. However, microbial-explicit models are diverse in terms of pool structure, representations of carbon flux dynamics, and environmental drivers, as illustrated in Figure 1.
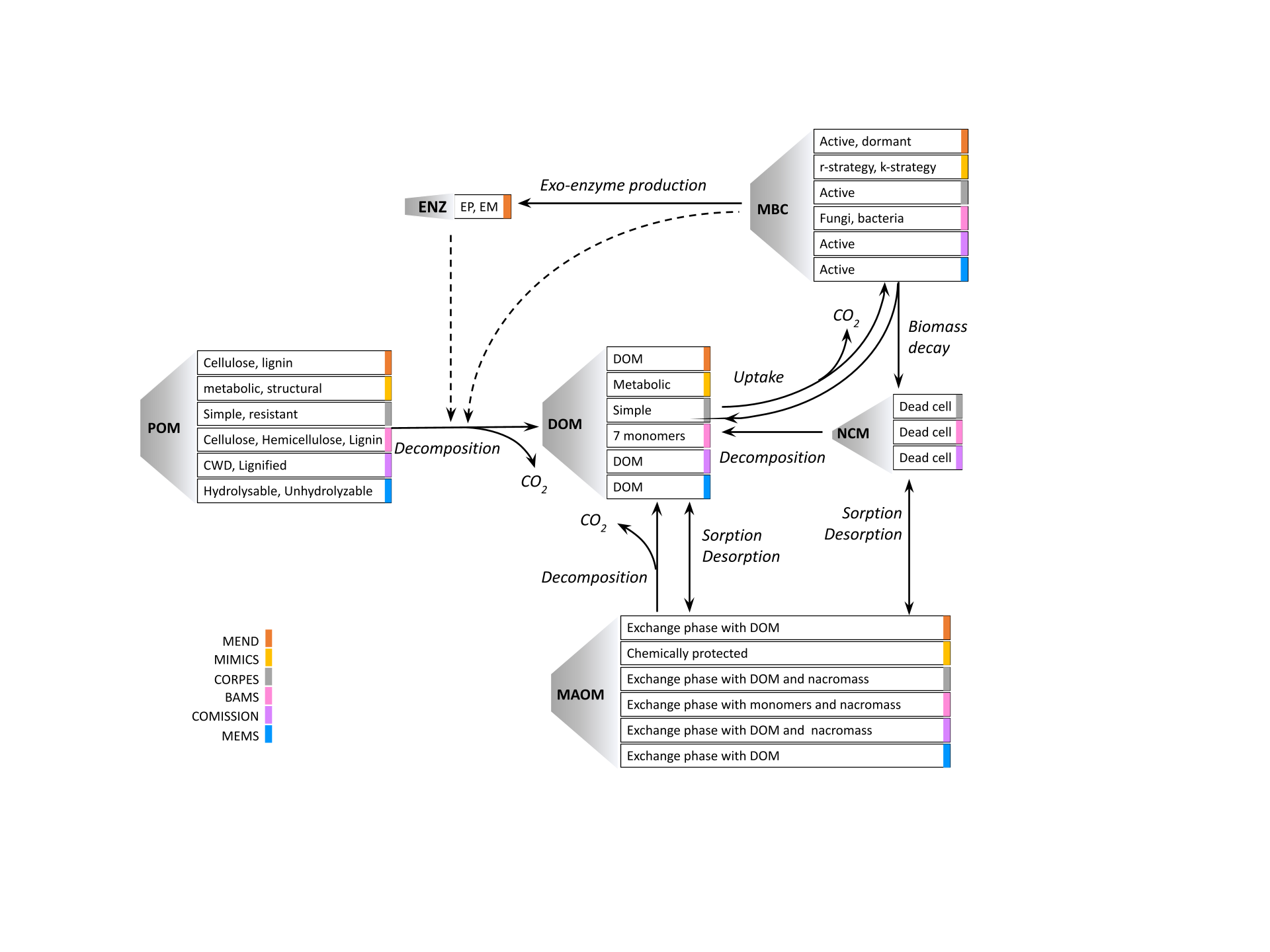
The variations in carbon model structures and dynamics contribute to
diverging long-term model projections, making model inter-comparisons
difficult. In order to explore, attribute, and better understand the
implications of varying the representations of the C dynamics, the
Microbial Explicit Model Consortium (MEMC) adopts a uniform pool
structure to enable easy model comparison exercises. By implementing
MEMC as an R package, we hope to make soil carbon modeling accessible to
a wider audience.
Model Structure
MEMC uses the common model structure visualized in Figure 2. In the first version of this package, users have the ability to decide which implementation POM decomposition (1), DOM uptake (2), and microbial biomass decay (3) as indicated in (Figure 3). This flexible framework allows users to isolate the effects of flux dynamics on model behavior.
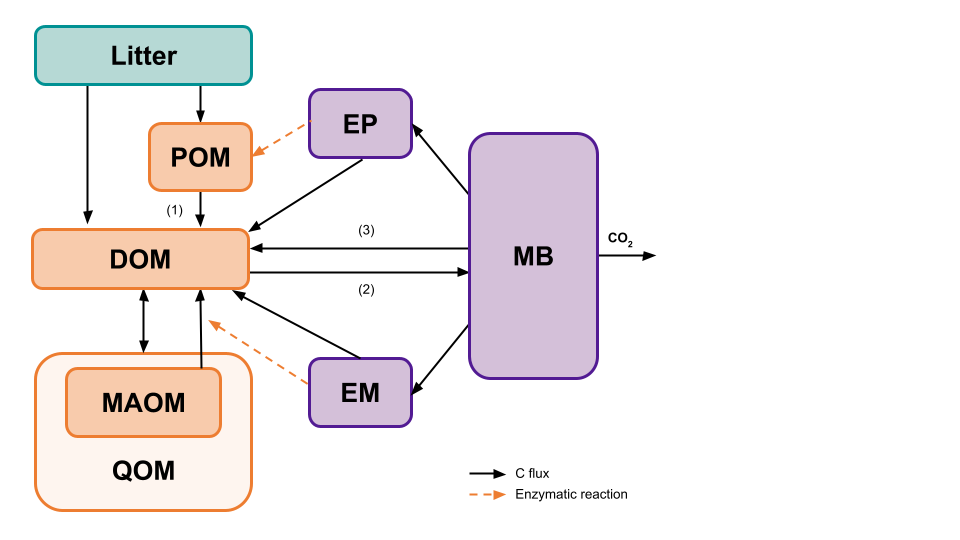
Model Configurations
The MEMC R package ships with six model pre-configured models (MICMS, MEND, CORPSE, BAMS, COMISSION, and MEMS). All of these model implementations use the uniform pool structure from Figure 2 but use a different combination of C flux dynamics as indicated in Figure 3.
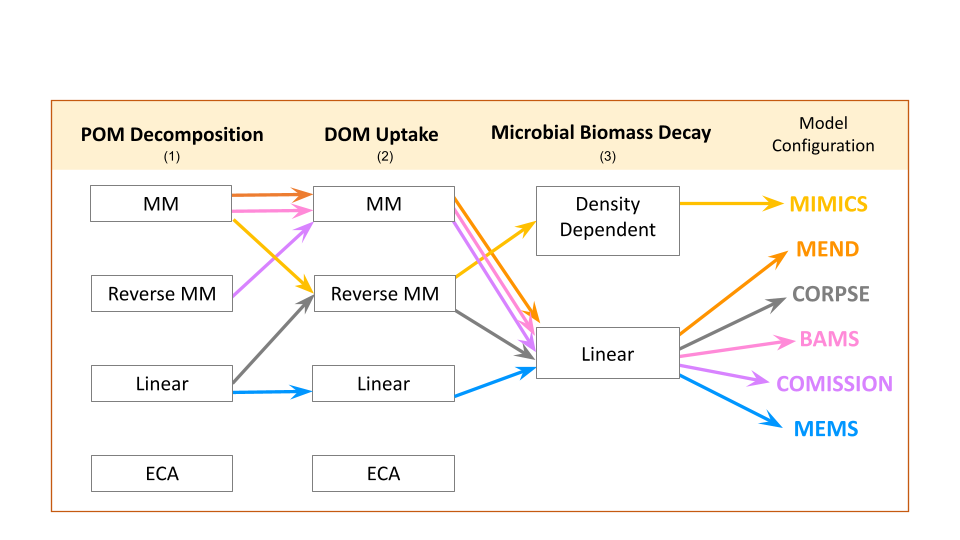
Dynamics
- MM: Michaelis Menten model, a commonly used equation in biogeochemical modelings which describe the rate of a reaction in response to a change in substrate.
- Reverse MM: Reverse Michaelis Menten model (RMM), a modified version of Michaelis Menten model, however now the modeled reaction rate changes varies in response to enzyme availability.
- Linear: a simple linear model (LM) is used.
-
Density Dependent: in the Density Dependent (DD)
model the density of the population (the microbial biomass) affects the
rate of a given process.
- ECA: Equilibrium Chemistry Approximation kinetics an alternative representation of biogeochemical processes that is more appropriate over a wide range of substrate availability.